Usefulness of measuring microRNAs in bile and plasma for pancreatic ductal adenocarcinoma (PDAC) diagnosis
Le Large TYS, Frampton AE, Meijer L, Stebbing J, Kazemier G, Giovannetti E.
Am J Gastroenterol. 2015 May;110(5):768-9. doi: 10.1038/ajg.2015.77
Abstract
We have read with interest the article by Cote and colleagues evaluating a microRNA (miRNA) signature panel in bile and plasma to distinguish PDAC from chronic pancreatitis (CP) and patients without pancreatic disease (1). These data add to the evidence for the feasibility of using cell-free miRNAs in biofluids for tumor screening. A plasma- and bile-panel of 5 miRNAs (miR-10b/-30c/-106b/-155/-212) emerged as a novel diagnostic test for PDAC, supporting future comparative studies with CA 19-9 and analyses of its’ prognostic value. Some key points should be discussed in more detail.
First, the selection of miRNAs on the basis of their overexpression in PDAC may not reflect the best discriminative cell-free miRNAs, since the mechanisms regulating miRNA release in the bloodstream and bile have not yet been clarified and several miRNAs have differential profiles in biofluids and tissues (2). This problem could have been avoided using a discovery microarray to profile biofluids from PDAC and CP. The same approach would have identified the best normalizer, since there is much debate on which endogenous miRNA can be used in biofluids (3). MiR-425-5p was selected because of the lowest cycle-threshold difference across samples amongst 4 candidate miRNAs. A microarray would have clarified the ideal normalizer for future routine use for measuring cell-free miRNAs in PDAC patients.
Second, the authors did not correlate miRNA levels to the histopathological features of the pancreatic tissues. Indeed, premalignant lesions like IPMNs or PanINs have overlapping upregulated miRNAs similar to PDAC, including for example miR-21 (4). This might explain the negative results concerning the diagnostic value of this miRNA, which could have been relatively high in CP and controls due to unknown premalignant lesions, or even due to the confounding effect of ductal obstruction or inflammation.
Importantly, a publically available dataset (GSE24279) showed that only 2 out of the 5 selected miRNAs relate to a malignant, rather than an inflammatory process (Fig.1).
Finally, we agree with the authors that a miRNA panel could also distinguish PDAC from other peri-ampullary malignancies. These tumors have similar symptoms and differential diagnosis is of utmost importance in the neoadjuvant era (5). However, this has never been shown in a comparative study in plasma or other biofluids.
In conclusion, we are indebted to Cote and colleagues for their study, and look forward to additional studies that may strengthen these results beyond already available PDAC biomarkers, as well as for prognostic purposes.
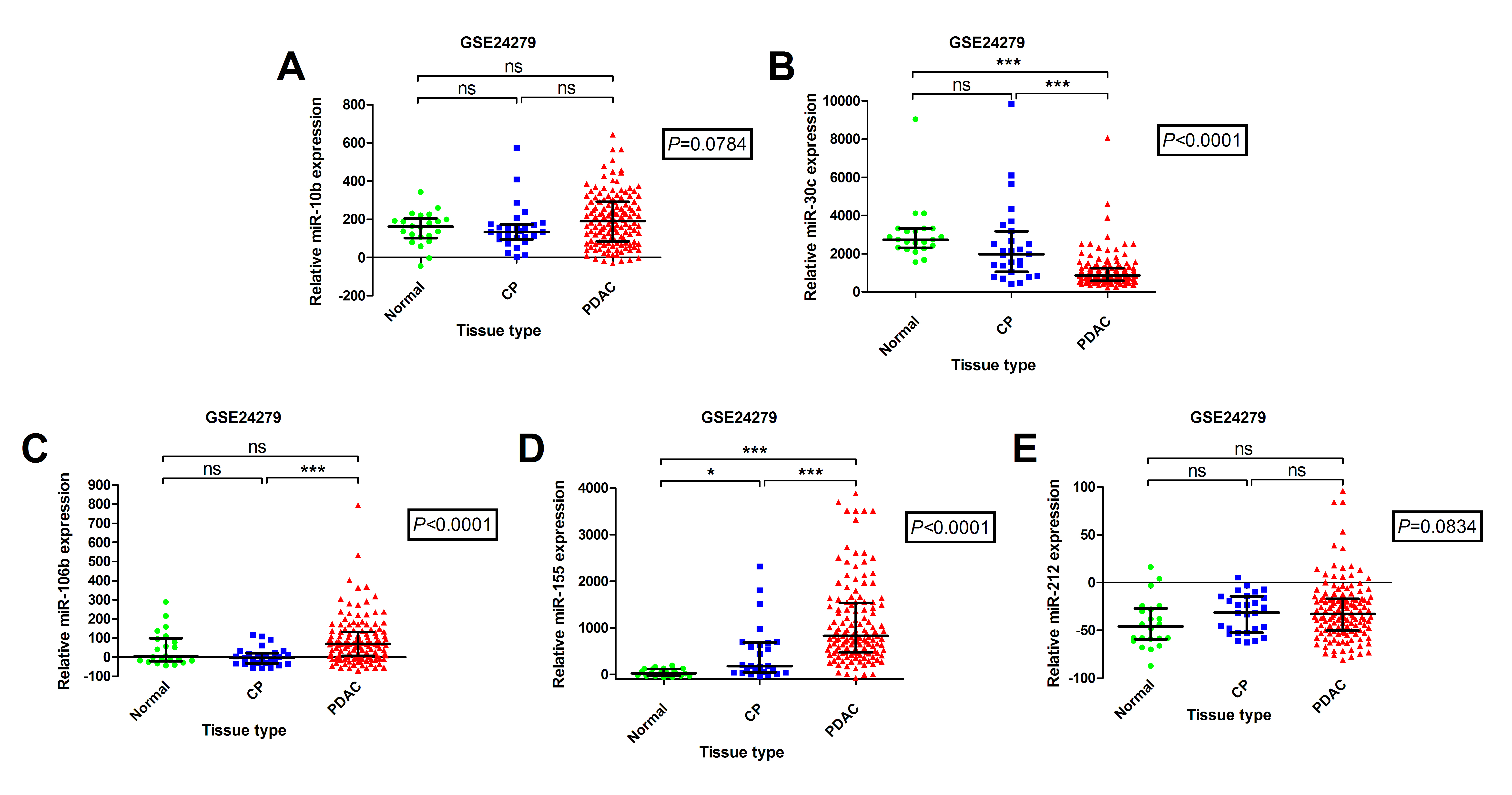 Down-regulated miR-30c and up-regulated miR-155 expression can differentiate PDAC tissues from normal pancreas and chronic pancreatitis. Using a publically available dataset (GSE24279) we examined (A) miR-10b; (B) miR-30c; (C) miR-106b; (D) miR-155 and (E) miR-212 expression in a large cohort of PDAC patients and compared it to chronic pancreatitis (CP) and normal pancreas (NP) samples. This dataset contains miRNA expression profiling results from tissue samples: PDAC (n = 136), CP (n=27) and NP (n=22). Due to an approximate normal distribution, the non-parametric Kruskal–Wallis one-way analysis of variance (ANOVA) was used to compare miRNA levels between tissue-types (P value in box), followed by Dunn’s multiple comparison test. Patients included in this study were of stages II, III and IV. Scatterplots are shown for each miRNA and the horizontal line represents the median expression level and inter-quartile range (***P<0.001; * P<0.05; ns, non-significant).
Down-regulated miR-30c and up-regulated miR-155 expression can differentiate PDAC tissues from normal pancreas and chronic pancreatitis. Using a publically available dataset (GSE24279) we examined (A) miR-10b; (B) miR-30c; (C) miR-106b; (D) miR-155 and (E) miR-212 expression in a large cohort of PDAC patients and compared it to chronic pancreatitis (CP) and normal pancreas (NP) samples. This dataset contains miRNA expression profiling results from tissue samples: PDAC (n = 136), CP (n=27) and NP (n=22). Due to an approximate normal distribution, the non-parametric Kruskal–Wallis one-way analysis of variance (ANOVA) was used to compare miRNA levels between tissue-types (P value in box), followed by Dunn’s multiple comparison test. Patients included in this study were of stages II, III and IV. Scatterplots are shown for each miRNA and the horizontal line represents the median expression level and inter-quartile range (***P<0.001; * P<0.05; ns, non-significant).
Link: www.ncbi.nlm.nih.gov/pubmed/25942302




